Michael Beer improves methodology for predicting disease-enabling genetic mutations
Researchers around the world have sequenced the genomes of patients suffering from common multigene diseases, looking for common mutations in their control regions. However these studies produce hundreds of mutations, and many of those prove to be benign.
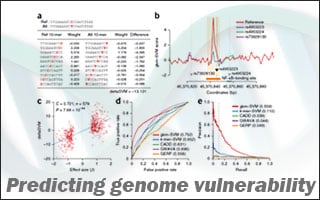
Now a research team led by Associate Professor of Biomedical Engineering Michael Beer have generated a new computational formula for predicting which mutations in control regions will wreak the most havoc. Their research was published in Nature Genetics.
Their focus was finding genetic control regions in stretches of DNA positioned near most genes. These regions act like dimmer switches that control the amount of each protein produced. Mutations in control regions generally have more subtle effects than mutations in genes themselves, but they can contribute to common, genetically complex chronic diseases such as diabetes.
Beer’s team first “trained” their computer program to recognize and measure DNase sensitivity in susceptible genetic control regions. DNase is an enzyme that cuts DNA wherever it is not tightly wound. The openness of particular sequences of DNA varies among different types of cells, and only control regions with open DNA can be active. How vulnerable certain stretches of DNA are to DNase is therefore an indication of which control regions are important in a given cell type.
They then computationally simulated “mutating” every DNA letter in turn and recalculated each section’s contribution to DNAse sensitivity. The larger the change in sensitivity after a given mutation, the more likely it is the that mutation will affect gene activity levels in the cell, Beer says.
The team compared these computer predictions to the known effects of some mutations and to the predictions made by alternative programs. When the programs’ “rules” were set to be equally thorough in their searches, Beer’s program was 56 percent accurate — 10 times more accurate than the next best program.
Beer worked with Andrew McCallion, PhD, an associate professor at the McKusick-Nathans Institute of Genetic Medicine at the Johns Hopkins University School of Medicine to further test the computational formula to predict the impact of mutations in the control regions for two pigment-related genes in mouse skin cells. They found that there was a strong correlation between the program’s prediction and the actual change experienced by the cells.
Dr. Beer and his team also tested their formula in mouse and human liver cells and in human leukemia cells and got similar results. They also tested their formula on three control region mutations already known to affect cholesterol levels, hemoglobin levels, and prostate cancer. Again they found that these mutations drew higher computer scores than other mutations in the same control regions.
Finally, the team examined the control regions for T helper cells, a type of immune cell that can contribute to autoimmune diseases. Their calculations identified 15 different control region mutations associated with nine different immune system disorders from allergies to multiple sclerosis and Crohn’s disease. The research honed in on the exact genetic mutation that mattered.
“The next step is to test gene activity levels in patients and find out if our predictions were right,” Beer says. “If so, it should help us determine how the activity is being perturbed and how we can fix it.”
This computational analysis can be repeated on many other diseases to provide timesaving insights for each.
Other authors of the report include Dongwon Lee, David Gorkin and Maggie Baker of the Johns Hopkins University School of Medicine and Benjamin Strober and Alessandro Asoni, who contributed to the project while undergraduates in the Department of Biomedical Engineering.
