Two studies led by Johns Hopkins Kimmel Cancer Center, Ludwig Center, and Johns Hopkins Whiting School of Engineering researchers report on a powerful new method that significantly improves the reliability and accuracy of artificial intelligence (AI) for many applications. As an example, they apply the new method to early cancer detection from blood samples, known as liquid biopsy.
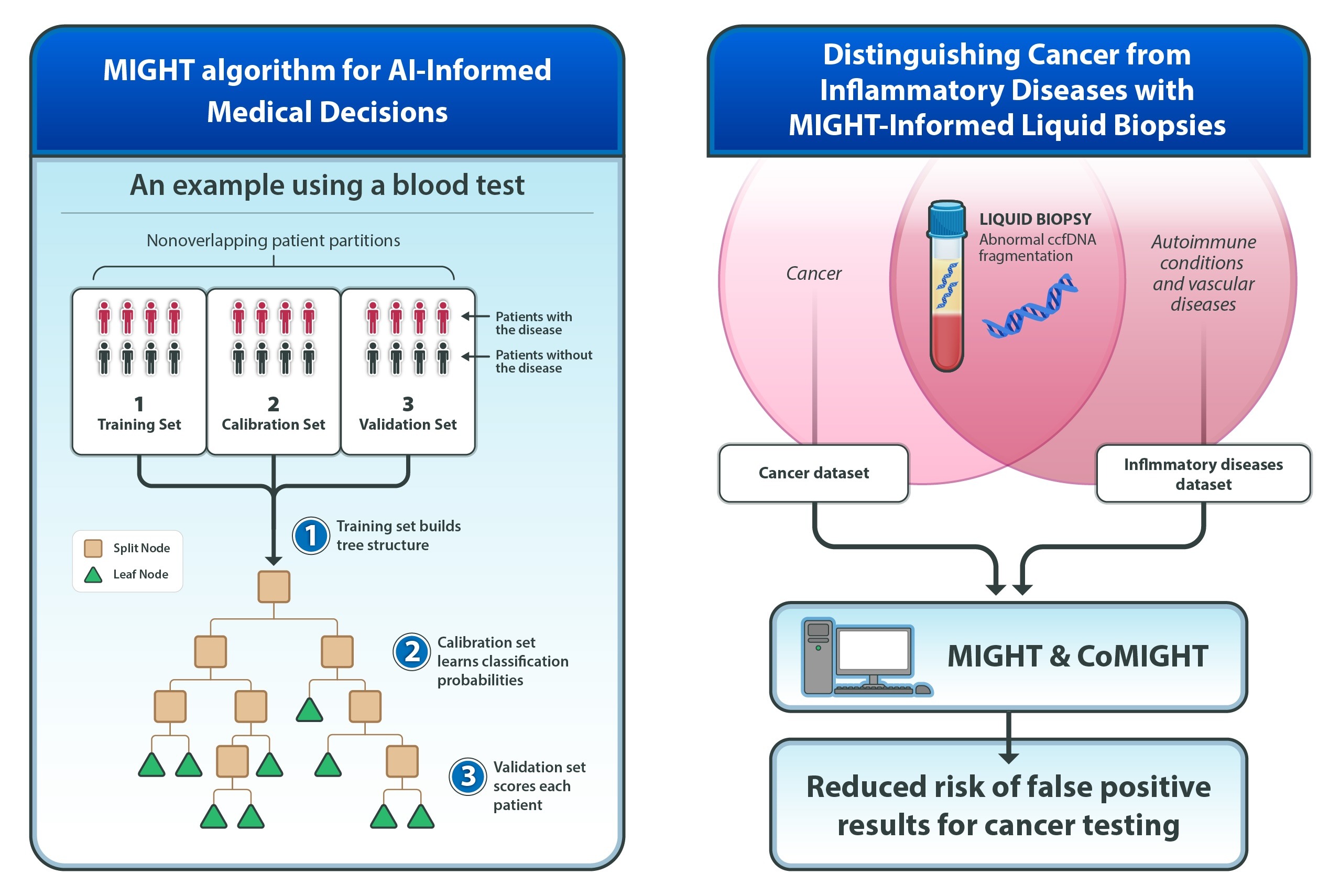
One study reports on the development of MIGHT (Multidimensional Informed Generalized Hypothesis Testing), an AI method that the researchers created to meet the high level of confidence needed for AI tools used in clinical decision making. To illustrate the benefits of MIGHT, they used it to develop a test for early cancer detection using circulating cell-free DNA (ccfDNA)—fragments of DNA circulating in the blood. A companion study found that ccfDNA fragmentation patterns used to detect cancer also appear in patients with autoimmune and vascular diseases. To develop a test with high sensitivity for cancer but reduced false-positive results, MIGHT was expanded to incorporate data from autoimmune and vascular diseases obtained from colleagues at Johns Hopkins and other institutions who treat and study these diseases.
The studies, supported in part by the National Institutes of Health, were published on Aug. 20 in the Proceedings of the National Academy of Sciences.
A related article, authored by three researchers from Johns Hopkins, Pixar co-founder Ed Catmull, Ph.D., and Microsoft chief data scientist of the AI for Good Lab Juan Lavista Ferres, was published concurrently in Cancer Discovery, a publication of the American Association for Cancer Research. It discusses the challenges of incorporating AI into clinical practice, including challenges addressed by MIGHT.
MIGHT fine-tunes itself using real data and checks its accuracy on different subsets of the data, using tens of thousands of decision-trees, and can be applied to any field employing big data, ranging from astronomy to zoology. It is particularly effective for the analysis of biomedical datasets with many variables but relatively few patient samples, a common situation in which traditional AI models often falter.
In tests using patient data, MIGHT consistently outperformed other AI methods in both sensitivity and consistency. It was applied to the blood of 1,000 individuals—352 patients with advanced cancers and 648 individuals without cancer. For each sample, the researchers evaluated 44 different variable sets, each consisting of a set of biological features, such as DNA fragment lengths or chromosomal abnormalities, and found that aneuploidy-based features (an abnormal number of chromosomes) delivered the best cancer detection performance with a sensitivity of 72% (ability to detect cancer) at 98% specificity (correctly identified those who were cancer free). This balance is critical in real-world medical applications where minimizing false positives is necessary to avoid unneeded procedures.
“MIGHT gives us a powerful way to measure uncertainty and increase reliability, especially in situations where sample sizes are limited but data complexity is high,” says Joshua Vogelstein, associate professor of biomedical engineering and a lead investigator.
MIGHT was also extended to a companion algorithm, called CoMIGHT, to determine whether combining multiple variable sets could improve cancer detection.