The Neural Signals and Computation lab is undertaking a number of efforts spanning data processing, modeling, and theory of how data can be used to elucidate biological understanding. On the data side, these projects include new data processing methods for high-dimensional neural recordings, such as multi-photon microscopy (including volumetric imaging and validation tools), high-density electrophysiology (e.g., Neuropixels), and functional Ultrasound (fUS). On the modeling side, projects include the statistical modeling of neural spiking behavior, including neural variability and dynamical systems models. On the theoretical side the lab includes projects analyzing properties of recurrent network networks. The lab’s advances are underscored by the development of general statistical models and techniques for correlated/structured signals (e.g. new dynamic filtering methods and fMRI analysis). Previous work includes analysis of hyperspectral imagery, computational neural networks, and stochastic filtering.
Decomposed dynamical systems models of neural data
Model: Dynamical systems offers a mathematical foundation with which to describe data with strong temporal relationships that unfold through time. Two primary models that learn dynamical systems from data include 1) nonlinear models that are stationary, or highly uninterpretable (e.g., Recurrent Neural Networks) and 2) linear systems that can be non-stationary (e.g., switched linear systems), but limited in the expressibility per number of parameters. Our group works with decomposed linear dynamical systems, i.e., systems that can be linearly broken up into core dynamics that are independent from each other, and thus represent separate and interpretable pieces of the full system. The methods paper can be found here.
Science: We used our dLDS framework to explore the dynamics of the C. elegans nervous system. We found that dLDS reveals both abrupt and gradual changes in the neural dynamics. These internal changes were similar, but not identical, over bouts of the same behavioral correlates, demonstrating gradual adaptation. In particular adaptation occurred post-stimulation with oxygen. Finally, by providing instantaneous measures of inter-neuron connectivity, we were able to define contextual roles for neurons that currently do not have well-known functional roles-in particular interneurons. Our preprint is available here.
Tracking millions of synapses via cross-trained image enhancement
Synaptic strength represents the wiring underlying our neural dynamics. This substrate is not static, but changes over time-especially in response to stress, learning, reward, and other salient environmental conditions. Imaging synaptic strength in vivo is fraught with difficulty due to the sub-micron size, thus limiting prior studies to small areas of the brain. By mixing fast resonant scanning of genetically labeled synapses with sophisticated image enhancement algorithms, however, we have demonstrated the capability to simultaneously image hundreds-of-thousands to millions of individual synapses. Our image enhancement method is based on learning to resolve one-photon data from ex vivo imaged slices to match simultaneously imaged super-resolution Airy-scan data. Imaging in vivo however, enables us to track individual synapse strength over time, seeing how the wiring of the brain changes in response to different external changes. We show that even static home-cage mice have up to 50% of their synapses change significantly over time—a number that rises to over 70% in enriched environments. You can find our paper here.
Tracking neurons in high-density electrode recordings
Studying the circuit level mechanisms of learning, adaptation, and other intrinsically long-term changes in neural function, require tracking the activity of individual neurons over long time-scales. Tracking neurons in electrophysiology, however, is complicated by the minimal information available in the voltage waveform recorded for every unit. In collaboration with Dr. Tim Harris’ group, we have been developing novel methods to automatically track single units across multiple months by leveraging spatial information afforded by new high-density probes, in particular Neuropixels. Using matching metrics based on cluster localization, waveform matching, and Earth-mover’s distance-based registration, we demonstrate the ability to track neurons across up to 47 days with 86% accuracy. The full paper can be found here.
Functional Ultrasound data processing
Whole brain recordings provide insight into how the global computational infrastructure of the brain coordinates to give rise to behavior. Current methods that can image deep structures at scale—i.e., fMRI—are limited to recordings in immobilized animals. Functional Ultrasound (fUS) provides a unique opportunity to bring such capabilities into freely moving settings, such as what is needed to understand global brain dynamics during naturalistic foraging. Our group is working on the necessary signal processing and data modeling necessary to robustly analyze such data. Initial work has created a motion correction, denoising, and functional component discovery pipeline. Our paper is available here, and we provide a website to disseminate out methods here.
Neural Anatomy and Optical Microscopy (NAOMi) Simulation
Functional fluorescence microscopy has become a staple in measuring neural activity during behavior in rodents, birds and fish (and more recently primates!). Recent advances have created both novel optical set-ups (e.g. vTwINS for volumetric imaging) and more complex algorithms for demixing the neural activity from the recorded fluorescence videos, however methods to validate these improved methods are lacking. NAOMi seeks to use the extensive literature on neural anatomy and knowledge of cellular calcium dynamics to provide simulations of fluorescence data with full ground truth of neural activity for validation purposes.
Detecting and correcting false transients in calcium image analysis
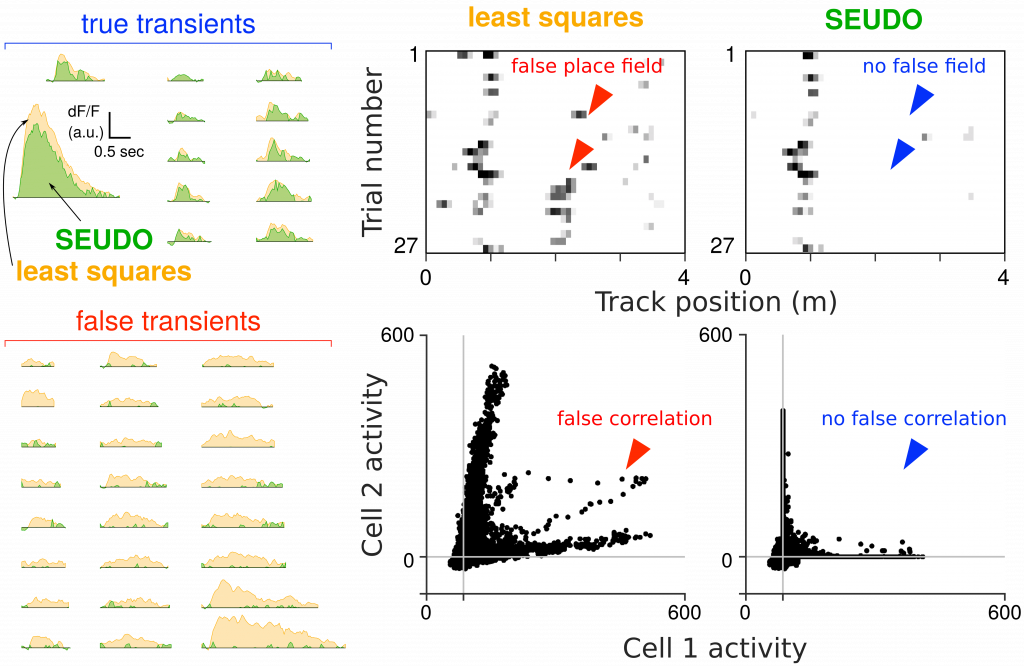
Accurate scientific results for circuits neuroscience using multi-photon calcium imaging depends heavily on accurate segmentation of the video into the neural activity for each individual neuron. Current automated time-trace extraction, regardless if whether sources were manually or automatically discovered, can erroneously attribute activity, resulting in scientifically significant errors such as spurious correlations or false representations (e.g. false place fields in the hippocampus). We devised methods to diagnose and correct such contaminating activity. To detect such errors we devised a combination of visualization of metrics via frequency-and-level of contamination (FaLCon) plots, as well as a user interface to easily permit practitioners to explore their data and identify falsely attributed activity. Furthermore, we also derived a new time-trace estimation algorithm robust to contamination from other sources. Our estimation procedure, Sparse Emulation of Unknown Dictionary Objects (SEUDO) directly models structured contaminants to the benefit of scientific discovery.
Two-Photon Microscopy
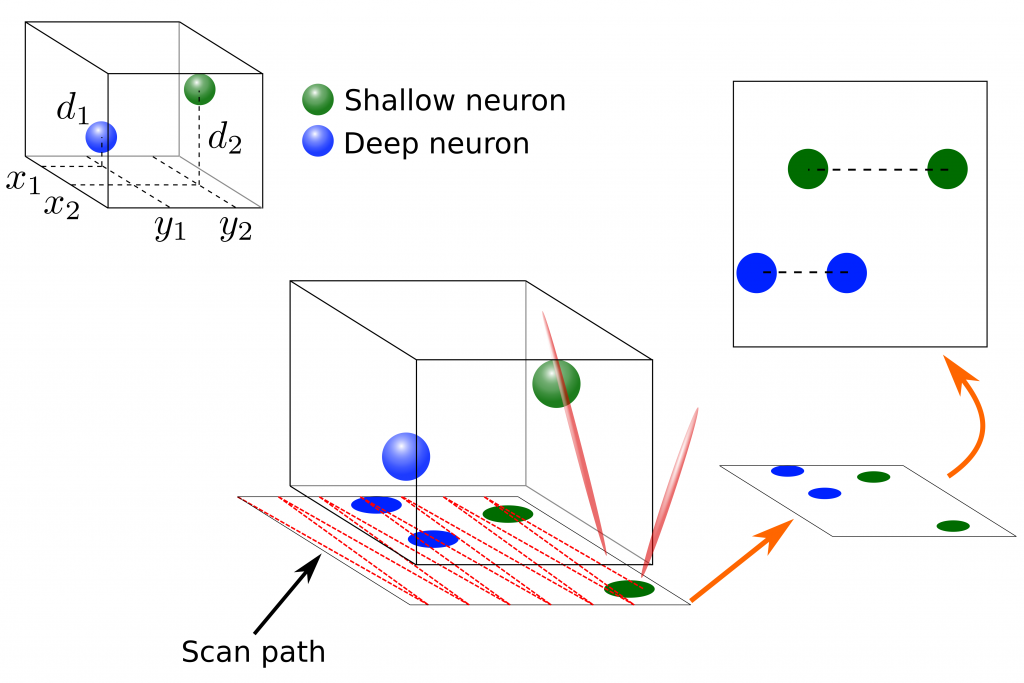
Two-Photon Microscopy (TPM) is a vital tool for recording large numbers of neurons over long time-scales. TPM recordings have allowed researchers to analyze entire networks of neurons in a number of cortical areas during awake behavior. Traditional TPM focuses on raster-scanning a single slice of neural tissue at relatively high frame-rates. To record additional neurons, volumetric imaging has been explored as well. Volumetric scanning, however, requires scanning many planes sequentially, lowering the overall scan rate. In this area I am collaborating with Dr. David Tank’s lab working on volumetric Two-photon Imaging of Neurons using Stereoscopy (vTwINS) method for imaging entire volumes with no reduction in frame rate. Specifically, our method records stereoscopic projections of the volume where each neuron is imaged twice, and the distance between the neuron’s images encodes the depth of that neuron. Our novel greedy demixing method, SCISM, can then decode these images and return the neural locations and activity patterns. In addition to volumetric imaging, I am also working on alternative pre-processing techniques to denoise TPM data and to robustly filter out structured fluorescence contaminations to get more accurate estimates of neural activity.
Statistical Modeling of Neural Spiking
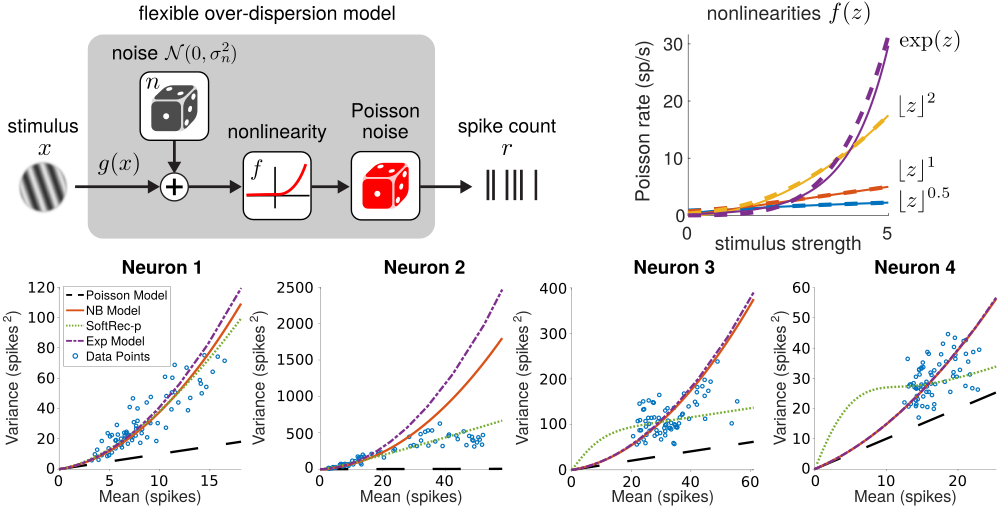
Interpreting neural spike trains is a critical step in decoding the relationship between neural activity and external stimuli. Basic probabilistic models of neural firing assume Poisson firing statistics. Recent work studying the over-dispersion of neural firing, however, has found that often the statistics of neural are decidedly not Poisson and may vary between neurons. Subsequent work has sought more flexible models which can account for the variability of neural firing. In this area I am working on creating simple yet flexible models for neural firing for which a small number of parameters can be learned directly (e.g. see code here) from neural recordings.
Matrix-normal Models of fMRI
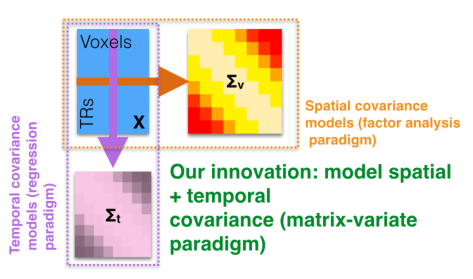
Functional Magnetic Resonance Imaging (fMRI) is a widely used modality for whole-brain imaging in humans. To better understand the cognitive processing taking place in the brain, many methods have been independently created to infer correlations between activity in different brain areas and across subjects and tasks. To further improve such inference I, with Michael Shvartsman and Mikio Aoi, have shown that the most prominent of these models can be placed in a single matrix-normal framework. This observation allows us to connect these models, devising faster, more flexible, and more accurate inference techniques for fMRI.
Analysis of Recurrent Neural Networks
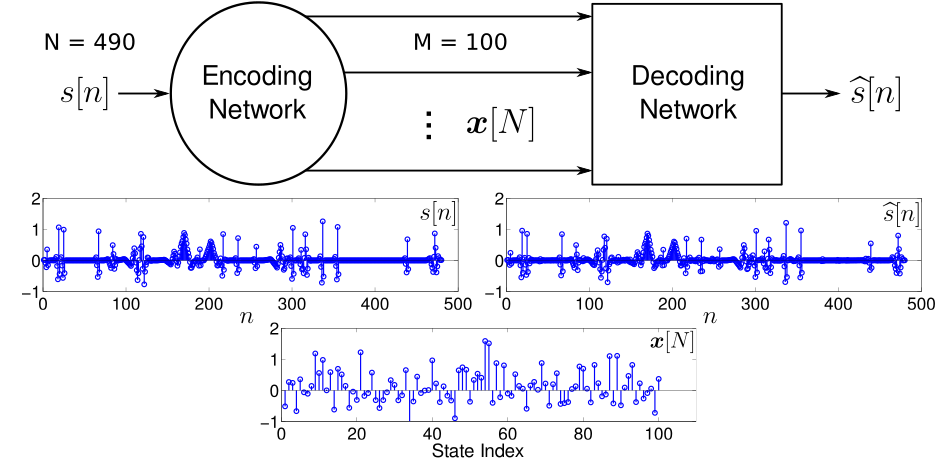
Interconnected networks of simple nodes have been shown to have computational abilities far beyond the sum of the individual neurons. Understanding how the network connectivity facilitates this huge increase in computational capacity has recently become increasingly important, in particular in the context of relating better understood theoretical network models and biological neural systems. In this area I’ve worked on both mathematical models of networks which compute the solution to various optimization problems, as well as deriving theoretical bounds on the short-term memory (STM) of linear neural networks. In particular I’ve worked on theory for single-input networks with sparse inputs and multiple-input networks with either sparse or low-rank inputs.
Sparsity-aware Stochastic Filtering
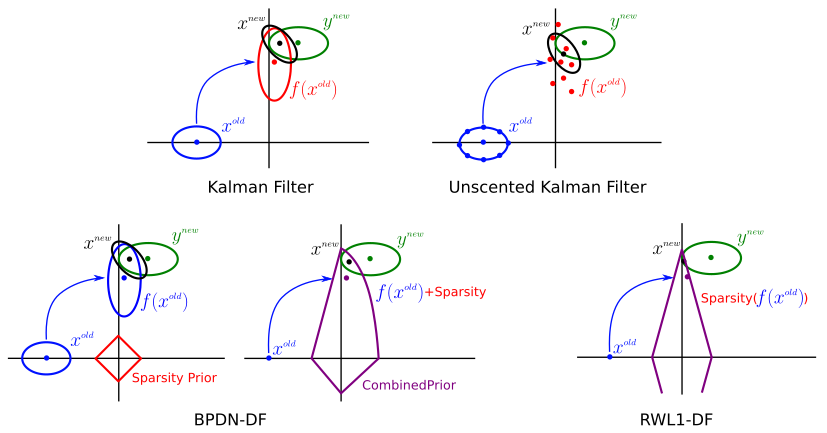
While the majority of work related to inferring and learning sparse signals and their representations has focused on static signals, (e.g., block processing of video), many applications with non-trivial temporal dynamics must be considered in a causal setting. I work to expand the ideas of sparse signal estimation to the realm of dynamic state tracking. The foundational formulation of the Kaman filter does not trivially expand to cover regimes of sparse signal and noise models, due to the loss of Gausian structure in the state statistics, as well as the impracticality of linear dynamics or retention of full covariance matrices. Instead other methods need to be developed. I am currently working on a series of algorithms based on probabilistic modeling to find fast updates for consecutive sparse state estimation. As an extension, similar methods can be employed to spatially correlated signals, resulting in more general, efficient multi-dimensional stochastic filtering techniques for correlated sparse signals.
Hyperspectral Imagery (HSI)
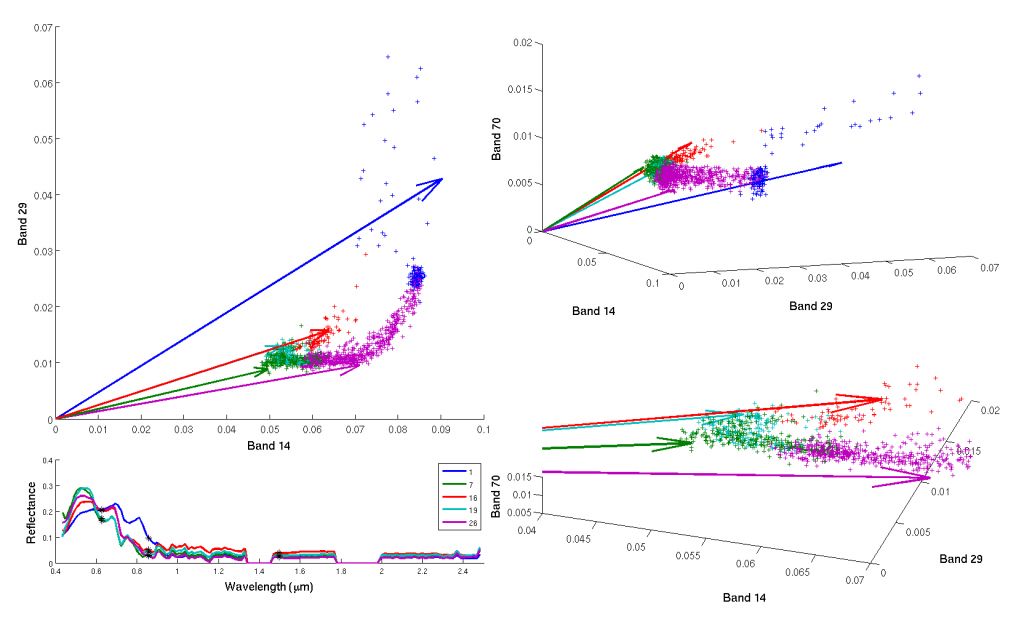
The sparse coding framework is particularly fitting for remote imaging using HSI. HSI uses many more spectral measurements than other imaging modalities (e.g., multispectral imaging (MSI) which typically takes ~8-12 spectral measurements), capturing data at 200-300+ wavelengths spanning the infrared to ultraviolet ranges. This level of spectral detail allows HSI to capture much richer information about the materials and features present in a scene. To discover the materials in a dataset, we can perform sparsity-based dictionary learning (code here). This unsupervised method extracts the specific spectra corresponding to different materials using only the basic assumption that few pure materials are present in any voxel. In addition to spectral demixing, I also use the sparsity-based inference procedures and learned dictionaries for unmixing, classification and other inverse problems that may arise in the use of HSI data. In particular, I have focused on spectral super-resolution of multispectral measurements to hyperspectral-level resolutions.